Introduction to Stability Selection
stabilityselectr.RmdThe stabilityselectr package performs stability
selection with a variety of kernels provided by the glmnet
package, and provides simple tools for plotting and extracting selected
features. There is additional functionality designed to facilitate
various forms of permutation clustering analyses.
Useful functions in stabilityselectr
stability_selection()is_stab_sel()get_stable_features()get_threshold_features()calc_emp_fdr()calc_emp_fdr_breaks()plot_emp_fdr()plot_permuted_data()progeny_cluster()stability_cluster()
Examples
A typical stability selection analysis might be similar to the one below.
Run stability_selection()
set.seed(101)
n_feat <- 20L
n_samples <- 100L
x <- matrix(rnorm(n_feat * n_samples), n_samples, n_feat)
colnames(x) <- paste0("feat", "_", head(letters, n_feat))
y <- sample(1:2, n_samples, replace = TRUE)
stab_sel <- stability_selection(x, y, kernel = "l1-logistic", num_iter = 500L)
#> ✓ Using kernel: 'l1-logistic' and 1 core (serial)
is_stab_sel(stab_sel)
#> [1] TRUE
stab_sel
#> ══ Stability Selection (Kernel: l1-logistic) ══════════════════════════
#> • Weakness (alpha) 0.8
#> • Weakness Probability (Pw) 0.5
#> • Number of Iterations 500
#> • Standardized 'Yes'
#> • Imputed Outliers 'No'
#> • Lambda Max 0.1879
#> • Lambda Min Ratio 0.1
#> • Permuted Data 'No'
#> • Random Seed 506
#> ═══════════════════════════════════════════════════════════════════════Stable Features at a threshold
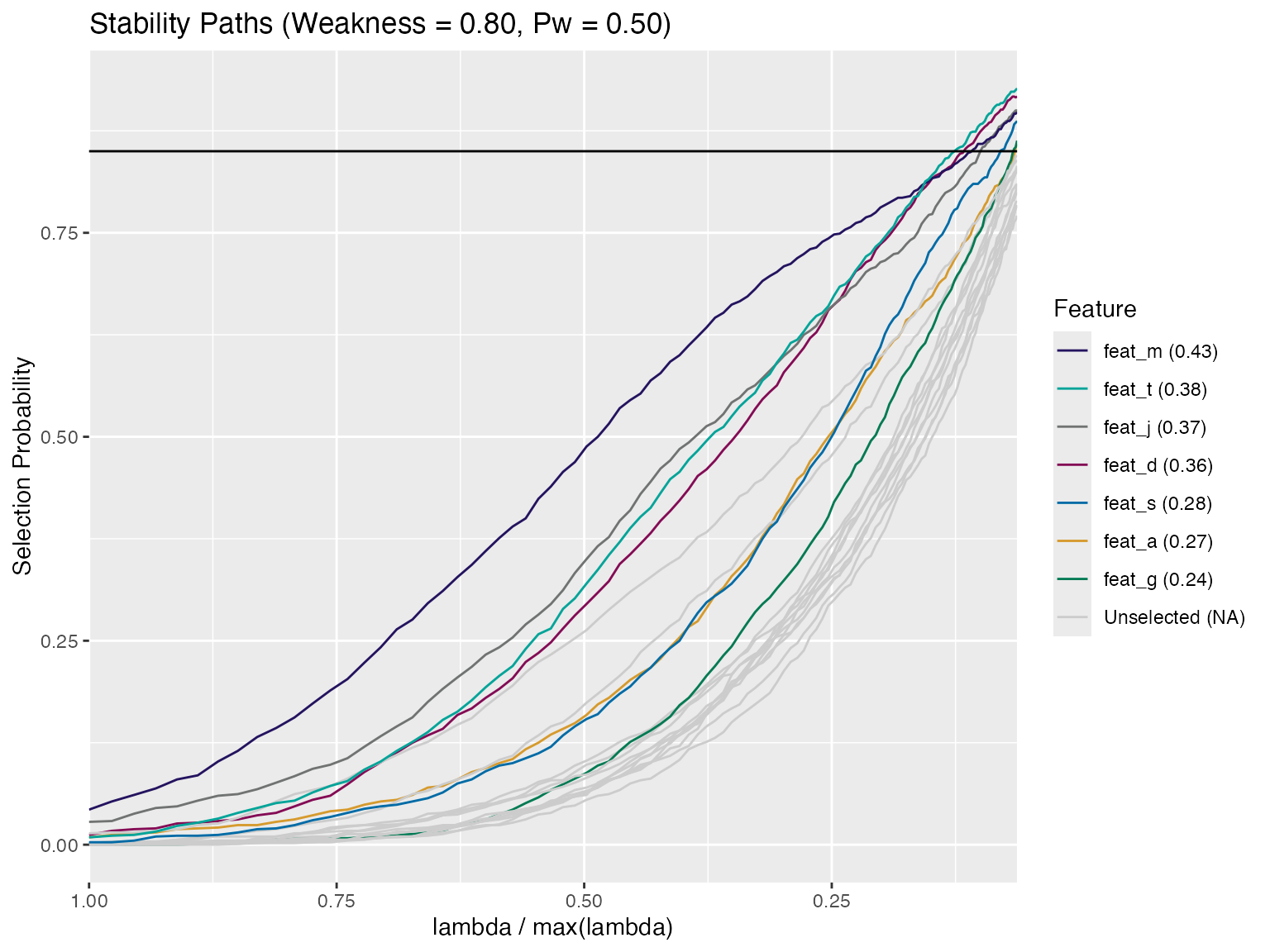
get_stable_features(stab_sel, thresh = 0.85)
#> MaxSelectProb FDRbound
#> feat_t 0.927 0.003571429
#> feat_d 0.917 0.007142857
#> feat_j 0.900 0.010714286
#> feat_m 0.898 0.014285714
#> feat_s 0.887 0.017857143
#> feat_g 0.863 0.021428571
#> feat_a 0.859 0.025000000Stable Features at multiple thresholds
get_threshold_features(stab_sel, thresh_vec = seq(0.7, 0.9, 0.05))
#> $thresh_0.7
#> MaxSelectProb FDRbound
#> feat_t 0.927 0.00625
#> feat_d 0.917 0.01250
#> feat_j 0.900 0.01875
#> feat_m 0.898 0.02500
#> feat_s 0.887 0.03125
#> feat_g 0.863 0.03750
#> feat_a 0.859 0.04375
#> feat_r 0.844 0.05000
#> feat_q 0.839 0.05625
#> feat_c 0.832 0.06250
#> feat_n 0.827 0.06875
#> feat_h 0.824 0.07500
#> feat_f 0.810 0.08125
#> feat_e 0.809 0.08750
#> feat_l 0.800 0.09375
#> feat_i 0.789 0.10000
#> feat_b 0.784 0.10625
#> feat_o 0.784 0.11250
#> feat_k 0.783 0.11875
#> feat_p 0.771 0.12500
#>
#> $thresh_0.75
#> MaxSelectProb FDRbound
#> feat_t 0.927 0.005
#> feat_d 0.917 0.010
#> feat_j 0.900 0.015
#> feat_m 0.898 0.020
#> feat_s 0.887 0.025
#> feat_g 0.863 0.030
#> feat_a 0.859 0.035
#> feat_r 0.844 0.040
#> feat_q 0.839 0.045
#> feat_c 0.832 0.050
#> feat_n 0.827 0.055
#> feat_h 0.824 0.060
#> feat_f 0.810 0.065
#> feat_e 0.809 0.070
#> feat_l 0.800 0.075
#> feat_i 0.789 0.080
#> feat_b 0.784 0.085
#> feat_o 0.784 0.090
#> feat_k 0.783 0.095
#> feat_p 0.771 0.100
#>
#> $thresh_0.8
#> MaxSelectProb FDRbound
#> feat_t 0.927 0.004166667
#> feat_d 0.917 0.008333333
#> feat_j 0.900 0.012500000
#> feat_m 0.898 0.016666667
#> feat_s 0.887 0.020833333
#> feat_g 0.863 0.025000000
#> feat_a 0.859 0.029166667
#> feat_r 0.844 0.033333333
#> feat_q 0.839 0.037500000
#> feat_c 0.832 0.041666667
#> feat_n 0.827 0.045833333
#> feat_h 0.824 0.050000000
#> feat_f 0.810 0.054166667
#> feat_e 0.809 0.058333333
#> feat_l 0.800 0.062500000
#>
#> $thresh_0.85
#> MaxSelectProb FDRbound
#> feat_t 0.927 0.003571429
#> feat_d 0.917 0.007142857
#> feat_j 0.900 0.010714286
#> feat_m 0.898 0.014285714
#> feat_s 0.887 0.017857143
#> feat_g 0.863 0.021428571
#> feat_a 0.859 0.025000000
#>
#> $thresh_0.9
#> MaxSelectProb FDRbound
#> feat_t 0.927 0.003125
#> feat_d 0.917 0.006250
#> feat_j 0.900 0.009375Progeny and Stability Clustering
See separate vignette on clustering:
vignette("progeny-clustering").