Introduction to helpr
helpr.RmdThe helpr package contains numerous helpers, wrappers,
and utilities used throughout the my analysis suite. It intentionally
favors base R over higher level tidyverse to minimize imports.
The goal is to provide an alternative gain in functionality without the
cost of additional imports/dependencies.
Useful functions in helpr
- File-system: See
?filesystem - Signal UI: See
?signal - Symbols: See
?sybml - Strings:
- Handlers:
-
liter():- List iteration, similar to
purrr::imap()but simpler
- List iteration, similar to
- Calculations:
-
calc_ccc(): Calculate Lin’s Concordance Correlation Coefficient for two vectors of numeric data. -
calc_ss(): Calculate the sum of squared errors: for numeric data.
-
- Logic Tests:
-
rep_lgl(): Are all the elements of a vector identical? -
is_monotonic(): Are the numerical elements of a vector monotonically increasing or decreasing? -
is_logspace(): Does a given object (vector,data.frame,tbl_df) appear to be in log-space? Note: this function is biased to proteomic data and should not be expected to be accurate for other applications out-of-the-box.
-
-
cross_tab()- Create a contingency table of counts generated by cross-classifying.
-
diff_vecs()- Generate all diffs of two vectors of (typically character) data.
Returns both sided
setdiff,unionandintersect.
- Generate all diffs of two vectors of (typically character) data.
Returns both sided
-
dater()- Generate a standardized data in (by default)
YYYY-MM-DD.
- Generate a standardized data in (by default)
Examples
cross_tab()
You do not need to “quote” the passed arguments, unquoted strings are
fine and are parsed by NSE (non-standard evaluation). The
... can take on either one or two column names:
# 1 factor
cross_tab(mtcars, cyl) # unquoted string
#> cyl
#> 4 6 8 Sum
#> 11 7 14 32
cross_tab(mtcars, "cyl") # quoted string
#> cyl
#> 4 6 8 Sum
#> 11 7 14 32
var <- "cyl"
cross_tab(mtcars, var) # external variable
#> cyl
#> 4 6 8 Sum
#> 11 7 14 32
cross_tab(mtcars, cyl, gear) # 2 factors
#> gear
#> cyl 3 4 5 Sum
#> 4 1 8 2 11
#> 6 2 4 1 7
#> 8 12 0 2 14
#> Sum 15 12 5 32
cross_tab(mtcars, cyl, gear, am) # 3 factors
#> , , am = 0
#>
#> gear
#> cyl 3 4 5 Sum
#> 4 1 2 0 3
#> 6 2 2 0 4
#> 8 12 0 0 12
#> Sum 15 4 0 19
#>
#> , , am = 1
#>
#> gear
#> cyl 3 4 5 Sum
#> 4 0 6 2 8
#> 6 0 2 1 3
#> 8 0 0 2 2
#> Sum 0 8 5 13
#>
#> , , am = Sum
#>
#> gear
#> cyl 3 4 5 Sum
#> 4 1 8 2 11
#> 6 2 4 1 7
#> 8 12 0 2 14
#> Sum 15 12 5 32
calc_ccc()
Calculate the ccc for two numeric vectors (visualize by
concordance):
x <- rnorm(100, mean = 10, sd = 0.5)
y <- x + rnorm(100, sd = 0.1) # add random scatter
ccc <- calc_ccc(x, y)
plot(x, y, pch = 21, cex = 1.75, col = NA,
bg = rgb(red = 0, green = 0, blue = 0, alpha = 0.5), # black w alpha
main = sprintf("The CCC = %0.3f with significance of p = %0.3f",
ccc$rho.c, ccc$p.value)
)
abline(0, 1, lty = 2, col = "blue", lwd = 1.5)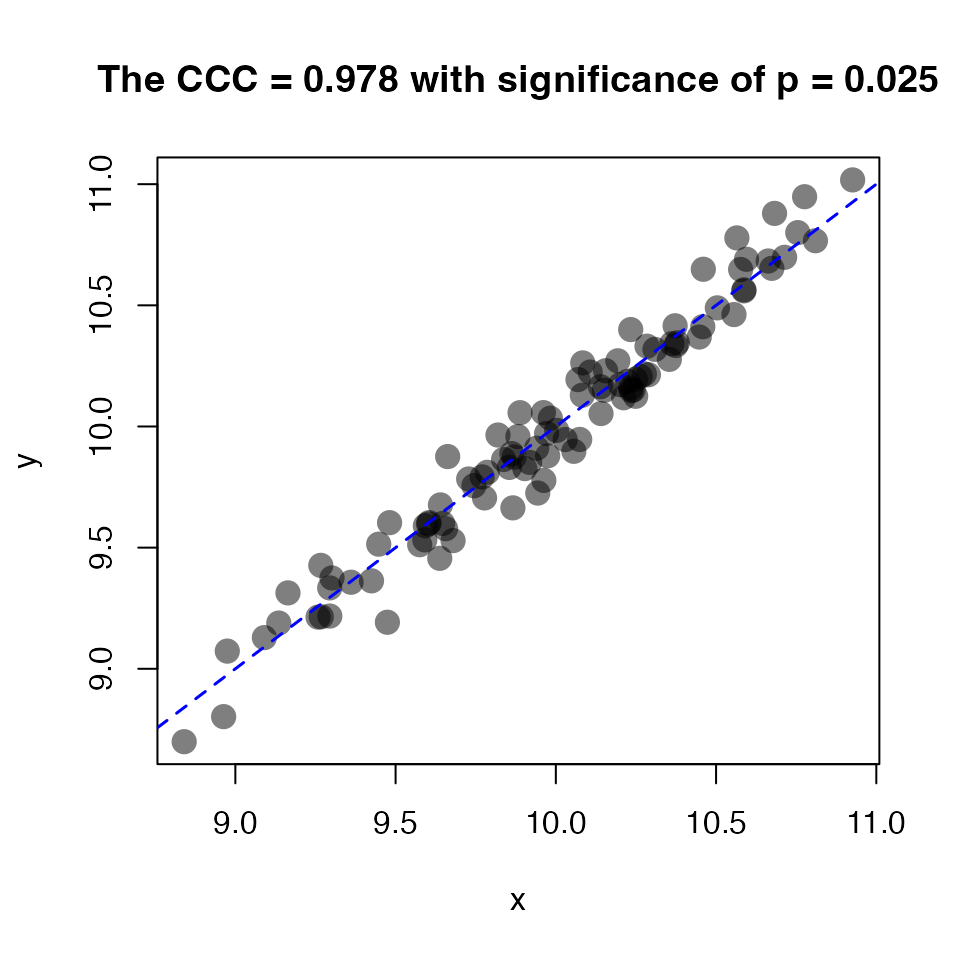
Concordance plot visualizing calc_ccc().
ccc
#> $rho_c
#> [1] 0.9778699
#>
#> $ci95
#> lower upper
#> 0.9675816 0.9849182
#>
#> $Z
#> [1] 2.24642
#>
#> $p_value
#> [1] 0.02467715
calc_ss()
Calculate the sum of squared differences for a numeric vector. Used ubiquotously in generating variances and standard deviations within other contexts (e.g. ANOVA, CVs):
is_monotonic()
is_monotonic(1:100)
#> [1] TRUE
is_monotonic(seq(-100, 100, by = 5)) # up
#> [1] TRUE
is_monotonic(seq(100, -100, by = -5)) # down
#> [1] TRUE
is_monotonic(rnorm(10))
#> [1] FALSE
is_logspace()
# A numeric vector
x <- rnorm(30, mean = 1000)
is_logspace(x)
#> [1] FALSE
is_logspace(log(x))
#> [1] TRUE
is_logspace(data) # FALSE
# log10-transform
for ( i in grep("^ft", names(data)) ) {
data[[i]] <- log10(data[[i]])
}
is_logspace(data) # base 10; TRUE
diff_vecs()
diff_vecs(LETTERS[1:10L], LETTERS[5:15L], verbose = TRUE) # return invisible
#> ℹ Vectors differ by:
#> • Unique to LETTERS[1:10L] >> 4
#> • Unique to LETTERS[5:15L] >> 5
#> • Common Intersect >> 6
#> • Union >> 15
(diff_vecs(LETTERS[1:10L], LETTERS[5:15L]))
#> $`unique_LETTERS[1:10L]`
#> [1] "A" "B" "C" "D"
#>
#> $`unique_LETTERS[5:15L]`
#> [1] "K" "L" "M" "N" "O"
#>
#> $inter
#> [1] "E" "F" "G" "H" "I" "J"
#>
#> $unique
#> [1] "A" "B" "C" "D" "E" "F" "G" "H" "I" "J" "K" "L" "M" "N" "O"